|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
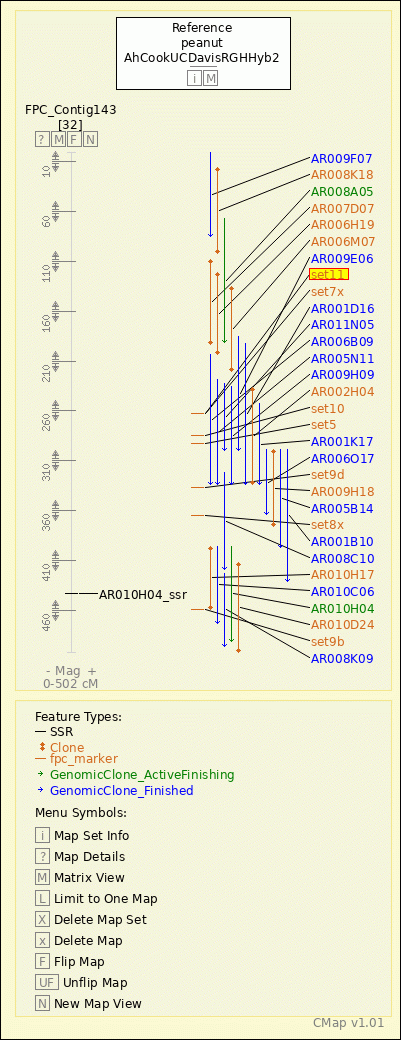
peanut - AhCookUCDavisRGHHyb2 - FPC_Contig143
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AR002H04
|
Clone
|
236 - 334
|
No other positions |
|
AR009F07
|
GenomicClone_Finished
|
0 - 85
|
No other positions |
|
AR006B09
|
GenomicClone_Finished
|
203 - 334
|
No other positions |
|
AR008A05
|
GenomicClone_ActiveFinishing
|
67 - 191
|
No other positions |
|
AR010H04
|
GenomicClone_ActiveFinishing
|
396 - 491
|
No other positions |
|
AR010H04_ssr
|
SSR
|
443.5
|
No other positions |
|
AR005B14
|
GenomicClone_Finished
|
299 - 397
|
No other positions |
|
AR010C06
|
GenomicClone_Finished
|
396 - 473
|
No other positions |
|
AR009H18
|
Clone
|
299 - 376
|
No other positions |
|
AR008K18
|
Clone
|
16 - 102
|
No other positions |
|
AR006O17
|
GenomicClone_Finished
|
299 - 364
|
No other positions |
|
AR008C10
|
GenomicClone_Finished
|
322 - 419
|
No other positions |
|
AR005N11
|
GenomicClone_Finished
|
228 - 334
|
No other positions |
|
AR006H19
|
Clone
|
121 - 203
|
No other positions |
|
AR008K09
|
GenomicClone_Finished
|
423 - 496
|
No other positions |
|
AR001D16
|
GenomicClone_Finished
|
192 - 334
|
No other positions |
|
AR009H09
|
GenomicClone_Finished
|
235 - 334
|
No other positions |
|
AR011N05
|
GenomicClone_Finished
|
232 - 300
|
No other positions |
|
AR001B10
|
GenomicClone_Finished
|
299 - 431
|
No other positions |
|
AR010H17
|
Clone
|
396 - 459
|
No other positions |
|
AR009E06
|
GenomicClone_Finished
|
185 - 300
|
No other positions |
|
AR007D07
|
Clone
|
108 - 193
|
No other positions |
|
AR001K17
|
GenomicClone_Finished
|
253 - 334
|
No other positions |
|
AR010D24
|
Clone
|
412 - 502
|
No other positions |
|
AR006M07
|
Clone
|
135 - 220
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial