|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
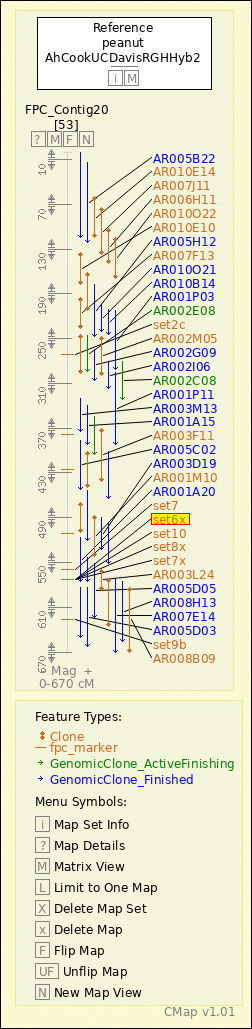
peanut - AhCookUCDavisRGHHyb2 - FPC_Contig20
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AR011I09
|
GenomicClone_Finished
|
0 - 115
|
No other positions |
|
AR003F11
|
Clone
|
370 - 442
|
No other positions |
|
AR010O22
|
Clone
|
115 - 169
|
No other positions |
|
AR005C02
|
GenomicClone_Finished
|
386 - 452
|
No other positions |
|
AR001D04
|
GenomicClone_Finished
|
401 - 464
|
No other positions |
|
AR002I06
|
GenomicClone_Finished
|
281 - 317
|
No other positions |
|
AR007E14
|
GenomicClone_Finished
|
573 - 670
|
No other positions |
|
AR008B09
|
Clone
|
584 - 670
|
No other positions |
|
AR002M05
|
Clone
|
247 - 299
|
No other positions |
|
AR002C08
|
GenomicClone_ActiveFinishing
|
281 - 331
|
No other positions |
|
AR001A15
|
GenomicClone_Finished
|
340 - 392
|
No other positions |
|
AR001M10
|
Clone
|
487 - 542
|
No other positions |
|
AR001A20
|
GenomicClone_Finished
|
517 - 575
|
No other positions |
|
AR010G04
|
Clone
|
161 - 212
|
No other positions |
|
AR010O21
|
GenomicClone_Finished
|
204 - 240
|
No other positions |
|
AR002A12
|
Clone
|
469 - 510
|
No other positions |
|
AR004P11
|
GenomicClone_Finished
|
498 - 552
|
No other positions |
|
AR007G15
|
GenomicClone_Finished
|
584 - 651
|
No other positions |
|
AR008H13
|
GenomicClone_Finished
|
576 - 656
|
No other positions |
|
AR002G09
|
GenomicClone_Finished
|
264 - 306
|
No other positions |
|
AR005D03
|
GenomicClone_Finished
|
584 - 663
|
No other positions |
|
AR002E08
|
GenomicClone_ActiveFinishing
|
246 - 294
|
No other positions |
|
AR001P03
|
GenomicClone_Finished
|
212 - 292
|
No other positions |
|
AR007J11
|
Clone
|
75 - 137
|
No other positions |
|
AR007F13
|
Clone
|
195 - 236
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial