|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
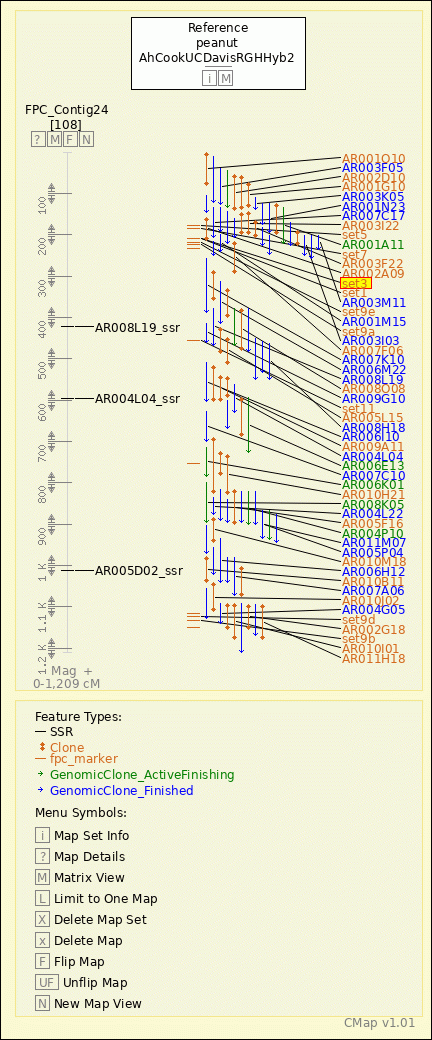
peanut - AhCookUCDavisRGHHyb2 - FPC_Contig24
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AR001M15
|
GenomicClone_Finished
|
200 - 241
|
No other positions |
|
AR001N23
|
GenomicClone_Finished
|
125 - 195
|
No other positions |
|
AR003F05
|
GenomicClone_Finished
|
41 - 128
|
No other positions |
|
AR007K10
|
GenomicClone_Finished
|
258 - 389
|
No other positions |
|
AR009G10
|
GenomicClone_Finished
|
415 - 460
|
No other positions |
|
AR006E19
|
GenomicClone_Finished
|
820 - 897
|
No other positions |
|
AR006H12
|
GenomicClone_Finished
|
935 - 1,041
|
No other positions |
|
AR006M01
|
GenomicClone_Finished
|
817 - 891
|
No other positions |
|
AR008H06
|
GenomicClone_ActiveFinishing
|
379 - 468
|
No other positions |
|
AR001O10
|
Clone
|
0 - 81
|
No other positions |
|
AR010C02
|
Clone
|
1,092 - 1,155
|
No other positions |
|
AR003I06
|
Clone
|
147 - 200
|
No other positions |
|
AR002G18
|
Clone
|
1,094 - 1,154
|
No other positions |
|
AR010J02
|
Clone
|
1,041 - 1,112
|
No other positions |
|
AR007F06
|
Clone
|
216 - 293
|
No other positions |
|
AR004L22
|
GenomicClone_Finished
|
835 - 877
|
No other positions |
|
AR004N14
|
GenomicClone_ActiveFinishing
|
866 - 937
|
No other positions |
|
AR005L15
|
Clone
|
448 - 514
|
No other positions |
|
AR010N05
|
GenomicClone_Finished
|
1,094 - 1,170
|
No other positions |
|
AR010B11
|
Clone
|
979 - 1,042
|
No other positions |
|
AR001B16
|
GenomicClone_Finished
|
1,092 - 1,209
|
No other positions |
|
AR001E19
|
GenomicClone_Finished
|
201 - 258
|
No other positions |
|
AR003K05
|
GenomicClone_Finished
|
110 - 140
|
No other positions |
|
AR007M09
|
GenomicClone_Finished
|
841 - 897
|
No other positions |
|
AR002D10
|
Clone
|
55 - 141
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial