|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
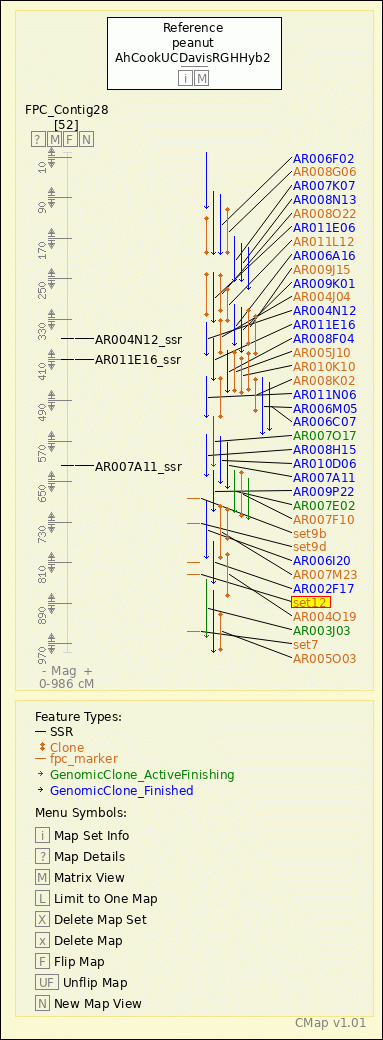
peanut - AhCookUCDavisRGHHyb2 - FPC_Contig28
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AR006I02
|
GenomicClone_Finished
|
77 - 202
|
No other positions |
|
AR011N06
|
GenomicClone_Finished
|
443 - 524
|
No other positions |
|
AR009J15
|
Clone
|
308 - 381
|
No other positions |
|
AR008O22
|
Clone
|
240 - 317
|
No other positions |
|
AR003J03
|
GenomicClone_ActiveFinishing
|
843 - 958
|
No other positions |
|
AR007G03
|
GenomicClone_ActiveFinishing
|
643 - 725
|
No other positions |
|
AR006I20
|
GenomicClone_Finished
|
690 - 801
|
No other positions |
|
AR008K02
|
Clone
|
445 - 513
|
No other positions |
|
AR009P22
|
GenomicClone_Finished
|
629 - 709
|
No other positions |
|
AR009K01
|
GenomicClone_Finished
|
307 - 395
|
No other positions |
|
AR004J04
|
Clone
|
328 - 400
|
No other positions |
|
AR007O17
|
GenomicClone_ActiveFinishing
|
522 - 620
|
No other positions |
|
AR008C16
|
GenomicClone_Finished
|
188 - 272
|
No other positions |
|
AR009B15
|
GenomicClone_Finished
|
0 - 112
|
No other positions |
|
AR005O03
|
Clone
|
908 - 985
|
No other positions |
|
AR001I08
|
Clone
|
443 - 525
|
No other positions |
|
AR006A16
|
GenomicClone_Finished
|
301 - 382
|
No other positions |
|
AR002F17
|
GenomicClone_Finished
|
768 - 850
|
No other positions |
|
AR008H15
|
GenomicClone_Finished
|
557 - 640
|
No other positions |
|
AR007M23
|
Clone
|
696 - 803
|
No other positions |
|
AR007F12
|
Clone
|
320 - 402
|
No other positions |
|
AR006M05
|
GenomicClone_Finished
|
445 - 557
|
No other positions |
|
AR011E16
|
GenomicClone_Finished
|
367 - 451
|
No other positions |
|
AR011E16_ssr
|
SSR
|
409
|
No other positions |
|
AR007K07
|
GenomicClone_Finished
|
167 - 256
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial