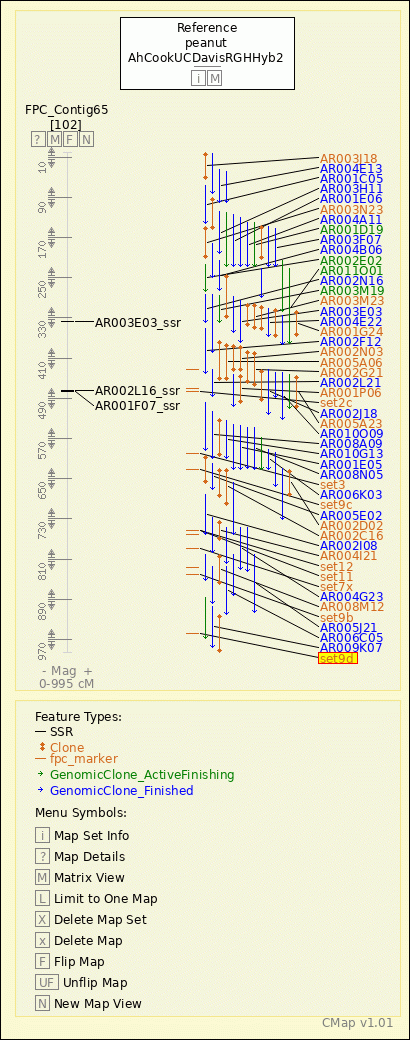
|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
peanut - AhCookUCDavisRGHHyb2 - FPC_Contig65
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AR002N16
|
GenomicClone_Finished
|
283 - 339
|
No other positions |
|
AR009L24
|
GenomicClone_Finished
|
498 - 594
|
No other positions |
|
AR002L21
|
GenomicClone_Finished
|
432 - 487
|
No other positions |
|
AR001D19
|
GenomicClone_ActiveFinishing
|
141 - 227
|
No other positions |
|
AR003M23
|
Clone
|
302 - 354
|
No other positions |
|
AR007G24
|
GenomicClone_Finished
|
800 - 883
|
No other positions |
|
AR006C05
|
GenomicClone_Finished
|
824 - 921
|
No other positions |
|
AR005K19
|
Clone
|
630 - 688
|
No other positions |
|
AR003B03
|
GenomicClone_Finished
|
287 - 360
|
No other positions |
|
AR005G18
|
GenomicClone_Finished
|
597 - 666
|
No other positions |
|
AR002I08
|
GenomicClone_Finished
|
682 - 762
|
No other positions |
|
AR005E02
|
GenomicClone_Finished
|
620 - 671
|
No other positions |
|
AR004E06
|
GenomicClone_Finished
|
800 - 853
|
No other positions |
|
AR002L16_ssr
|
SSR
|
474.5
|
No other positions |
|
AR003L07
|
GenomicClone_Finished
|
182 - 250
|
No other positions |
|
AR004E13
|
GenomicClone_Finished
|
34 - 100
|
No other positions |
|
AR005I01
|
Clone
|
395 - 461
|
No other positions |
|
AR011P12
|
Clone
|
432 - 496
|
No other positions |
|
AR002H06
|
Clone
|
244 - 332
|
No other positions |
|
AR002E02
|
GenomicClone_ActiveFinishing
|
224 - 277
|
No other positions |
|
AR006G21
|
GenomicClone_Finished
|
542 - 632
|
No other positions |
|
AR004A11
|
GenomicClone_Finished
|
140 - 227
|
No other positions |
|
AR005F15
|
GenomicClone_Finished
|
748 - 835
|
No other positions |
|
AR003E17
|
GenomicClone_Finished
|
439 - 489
|
No other positions |
|
AR001B20
|
GenomicClone_ActiveFinishing
|
215 - 318
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial