|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
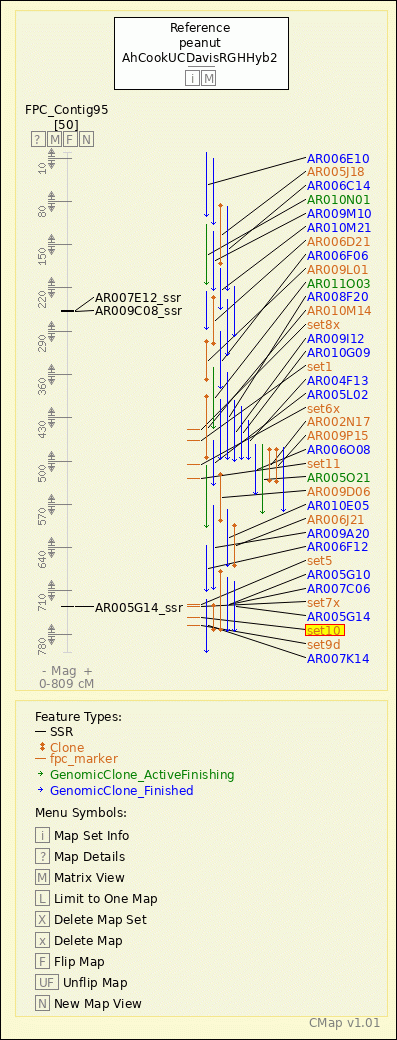
peanut - AhCookUCDavisRGHHyb2 - FPC_Contig95
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AR004F23
|
Clone
|
730 - 776
|
No other positions |
|
AR010M14
|
Clone
|
393 - 498
|
No other positions |
|
AR010G09
|
GenomicClone_Finished
|
411 - 498
|
No other positions |
|
AR001O17
|
GenomicClone_Finished
|
400 - 498
|
No other positions |
|
AR009D06
|
Clone
|
518 - 600
|
No other positions |
|
AR009P15
|
Clone
|
478 - 537
|
No other positions |
|
AR006O08
|
GenomicClone_Finished
|
474 - 554
|
No other positions |
|
AR006M11
|
GenomicClone_Finished
|
239 - 329
|
No other positions |
|
AR006J21
|
Clone
|
601 - 672
|
No other positions |
|
AR009L01
|
Clone
|
304 - 372
|
No other positions |
|
AR010E05
|
GenomicClone_Finished
|
578 - 670
|
No other positions |
|
AR010A12
|
GenomicClone_Finished
|
11 - 118
|
No other positions |
|
AR008F20
|
GenomicClone_Finished
|
357 - 498
|
No other positions |
|
AR006F06
|
GenomicClone_Finished
|
291 - 383
|
No other positions |
|
AR005J18
|
Clone
|
83 - 183
|
No other positions |
|
AR007C06
|
GenomicClone_Finished
|
688 - 776
|
No other positions |
|
AR009C08
|
GenomicClone_Finished
|
217 - 299
|
No other positions |
|
AR009C08_ssr
|
SSR
|
258
|
No other positions |
|
AR005G10
|
GenomicClone_Finished
|
688 - 776
|
No other positions |
|
AR004F13
|
GenomicClone_Finished
|
434 - 498
|
No other positions |
|
AR005G14
|
GenomicClone_Finished
|
695 - 776
|
No other positions |
|
AR005G14_ssr
|
SSR
|
735.5
|
No other positions |
|
AR005O21
|
GenomicClone_ActiveFinishing
|
474 - 585
|
No other positions |
|
AR010M21
|
GenomicClone_Finished
|
189 - 255
|
No other positions |
|
AR005L02
|
GenomicClone_Finished
|
466 - 539
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial