|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
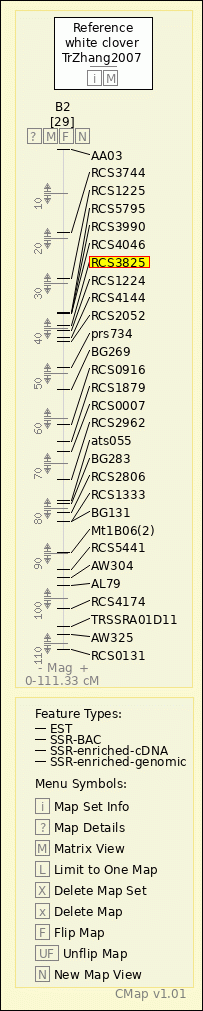
white clover - TrZhang2007 - B2
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AA03
|
EST
|
0
|
No other positions |
|
RCS3744
|
EST
|
18.49
|
white clover - TrZhang2007 - B1
|
RCS3744
|
EST
|
63.99 (cM)
|
Automated name-based
|
View Maps
|
|
RCS1225
|
SSR-enriched-cDNA
|
28.83
|
No other positions |
|
RCS5795
|
SSR-enriched-genomic
|
36.32
|
No other positions |
|
RCS3990
|
EST
|
36.57
|
No other positions |
|
RCS4046
|
EST
|
36.67
|
white clover - TrZhang2007 - B1
|
RCS4046
|
EST
|
56.72 (cM)
|
Automated name-based
|
View Maps
|
|
RCS3825
|
EST
|
39.35
|
white clover - TrZhang2007 - B1
|
RCS3825
|
EST
|
127.14 (cM)
|
Automated name-based
|
View Maps
|
|
RCS1224
|
SSR-enriched-cDNA
|
40.47
|
white clover - TrZhang2007 - C2
|
RCS1224
|
SSR-enriched-cDNA
|
9.21 (cM)
|
Automated name-based
|
View Maps
|
|
RCS4144
|
EST
|
41.97
|
No other positions |
|
RCS2052
|
SSR-enriched-genomic
|
42.86
|
No other positions |
|
prs734
|
EST
|
48.68
|
No other positions |
|
BG269
|
EST
|
53.5
|
No other positions |
|
RCS0916
|
SSR-enriched-genomic
|
61.33
|
No other positions |
|
RCS1879
|
SSR-enriched-cDNA
|
65.07
|
No other positions |
|
RCS0007
|
SSR-enriched-cDNA
|
67.35
|
No other positions |
|
RCS2962
|
SSR-enriched-cDNA
|
73.53
|
white clover - TrZhang2007 - G1
|
RCS2962
|
SSR-enriched-cDNA
|
30.52 (cM)
|
Automated name-based
|
View Maps
|
|
ats055
|
SSR-enriched-genomic
|
78.31
|
white clover - TrZhang2007 - D1
|
ats055
|
SSR-enriched-genomic
|
35.38 (cM)
|
Automated name-based
|
View Maps
|
|
BG283
|
EST
|
78.87
|
No other positions |
|
RCS2806
|
SSR-enriched-cDNA
|
80.91
|
white clover - TrZhang2007 - A2
|
RCS2806
|
SSR-enriched-cDNA
|
0 (cM)
|
Automated name-based
|
View Maps
|
|
RCS1333
|
SSR-enriched-cDNA
|
82.92
|
white clover - TrZhang2007 - A1
|
RCS1333
|
SSR-enriched-cDNA
|
52.35 (cM)
|
Automated name-based
|
View Maps
|
|
BG131
|
EST
|
83
|
No other positions |
|
Mt1B06(2)
|
SSR-BAC
|
89.93
|
No other positions |
|
RCS5441
|
EST
|
93.62
|
No other positions |
|
AW304
|
EST
|
95.33
|
No other positions |
|
AL79
|
EST
|
97.18
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial