|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
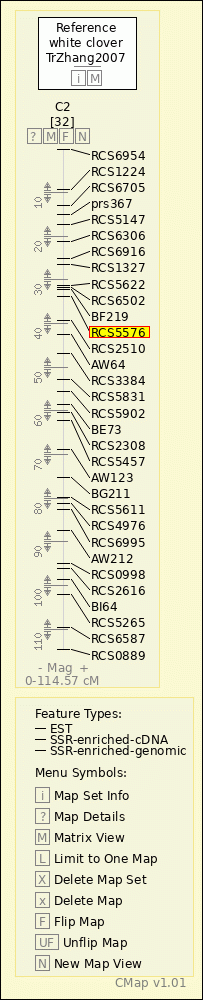
white clover - TrZhang2007 - C2
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
RCS6954
|
EST
|
0
|
white clover - TrZhang2007 - C1
|
RCS6954
|
EST
|
42.27 (cM)
|
Automated name-based
|
View Maps
|
|
RCS1224
|
SSR-enriched-cDNA
|
9.21
|
white clover - TrZhang2007 - B2
|
RCS1224
|
SSR-enriched-cDNA
|
40.47 (cM)
|
Automated name-based
|
View Maps
|
|
RCS6705
|
EST
|
13.01
|
No other positions |
|
prs367
|
EST
|
15
|
No other positions |
|
RCS5147
|
EST
|
17.19
|
No other positions |
|
RCS6306
|
EST
|
21.14
|
white clover - TrZhang2007 - C1
|
RCS6306
|
EST
|
8.9 (cM)
|
Automated name-based
|
View Maps
|
|
RCS6916
|
EST
|
25.01
|
white clover - TrZhang2007 - C1
|
RCS6916
|
EST
|
33.03 (cM)
|
Automated name-based
|
View Maps
|
|
RCS1327
|
SSR-enriched-cDNA
|
26.37
|
No other positions |
|
RCS5622
|
SSR-enriched-genomic
|
31.25
|
No other positions |
|
RCS6502
|
EST
|
31.68
|
No other positions |
|
BF219
|
EST
|
32.16
|
No other positions |
|
RCS5576
|
SSR-enriched-genomic
|
33.72
|
white clover - TrZhang2007 - E1
|
RCS5576
|
SSR-enriched-genomic
|
110.58 (cM)
|
Automated name-based
|
View Maps
|
|
RCS2510
|
SSR-enriched-cDNA
|
39.33
|
No other positions |
|
AW64
|
EST
|
42.42
|
No other positions |
|
RCS3384
|
SSR-enriched-cDNA
|
46.77
|
white clover - TrZhang2007 - H1
|
RCS3384
|
SSR-enriched-cDNA
|
63.86 (cM)
|
Automated name-based
|
View Maps
|
|
RCS5831
|
SSR-enriched-genomic
|
52.8
|
No other positions |
|
RCS5902
|
SSR-enriched-genomic
|
55.51
|
No other positions |
|
BE73
|
EST
|
58.63
|
white clover - TrZhang2007 - G1
|
BE73
|
EST
|
68.67 (cM)
|
Automated name-based
|
View Maps
|
|
white clover - TrZhang2007 - C1
|
BE73
|
EST
|
54.03 (cM)
|
Automated name-based
|
View Maps
|
|
RCS2308
|
SSR-enriched-genomic
|
60.32
|
No other positions |
|
RCS5457
|
SSR-enriched-genomic
|
62.26
|
No other positions |
|
AW123
|
EST
|
68.96
|
No other positions |
|
BG211
|
EST
|
75.19
|
No other positions |
|
RCS5611
|
SSR-enriched-genomic
|
79.85
|
No other positions |
|
RCS4976
|
EST
|
81.13
|
No other positions |
|
RCS6995
|
EST
|
82.68
|
white clover - TrZhang2007 - C1
|
RCS6995
|
EST
|
66.06 (cM)
|
Automated name-based
|
View Maps
|
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial