|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
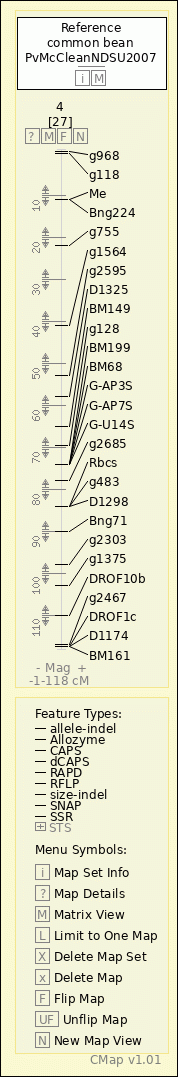
common bean - PvMcCleanNDSU2007 - 4
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
Me
|
Allozyme
|
0 - 22
|
No other positions |
|
g118
|
SSR
|
0
|
No other positions |
|
g968
|
dCAPS
|
-1 - 0
|
No other positions |
|
Bng224
|
RFLP
|
0 - 22
|
soybean - GmComposite2003 - K
|
Bng224_1
|
RFLP
|
75.23 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
common bean - BAT93_x_JALOEEP558_b - B04
|
Bng224
|
RFLP
|
14.07 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
g755
|
size-indel
|
22
|
No other positions |
|
g1564
|
CAPS
|
41
|
No other positions |
|
g2595
|
CAPS
|
53
|
No other positions |
|
D1325
|
RFLP
|
58
|
common bean - BAT93_x_JALOEEP558_b - B04
|
D1325
|
RFLP
|
19.35 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
BM149
|
SSR
|
65
|
common bean - PvConsensus_GaleanoFernandez2011_a - Pv04
|
BM149
|
SSR
|
99.16 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
g128
|
allele-indel
|
65 - 74
|
No other positions |
|
BM199
|
SSR
|
74
|
common bean - PvConsensus_GaleanoFernandez2011_a - Pv04
|
BM199
|
SSR
|
158.25 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
BM68
|
SSR
|
74
|
common bean - PvConsensus_GaleanoFernandez2011_a - Pv04
|
BM68
|
SSR
|
158.25 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
G-AP3S
|
STS
|
74
|
No other positions |
|
G-AP7S
|
STS
|
74
|
No other positions |
|
G-U14S
|
STS
|
74
|
No other positions |
|
g2685
|
size-indel
|
78
|
No other positions |
|
Rbcs
|
Allozyme
|
78 - 90
|
No other positions |
|
g483
|
dCAPS
|
78 - 90
|
No other positions |
|
D1298
|
RFLP
|
78 - 90
|
common bean - BAT93_x_JALOEEP558_b - B04
|
D1298
|
RFLP
|
60.17 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
Bng71
|
RFLP
|
90
|
No other positions |
|
g2303
|
SNAP
|
98
|
No other positions |
|
g1375
|
CAPS
|
103
|
No other positions |
|
DROF10b
|
RAPD
|
110
|
No other positions |
|
g2467
|
CAPS
|
117
|
No other positions |
|
DROF1c
|
RAPD
|
117 - 118
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial