|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
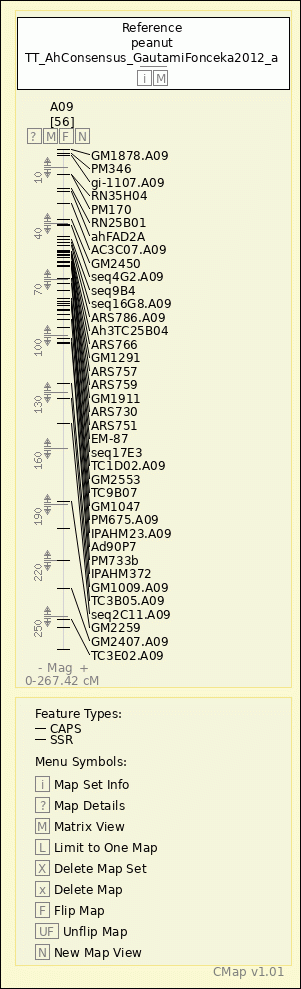
peanut - TT_AhConsensus_GautamiFonceka2012_a - A09
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
GM1878.A09
|
SSR
|
0
|
peanut - TT_ICGS44_x_ICGS76_a - LG12
|
GM1878
|
SSR
|
85.9 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TAG24_x_ICGV86031_a - LG_AhXIII
|
GM1878
|
SSR
|
204.6 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhComposite_SujayGowda2012_a - AhV
|
GM1878
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TG26_x_GPBD4_a - AhV
|
GM1878
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiPandey2012_a - LG_AhXIII
|
GM1878
|
SSR
|
192.59 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.duranensis_a - A05
|
GM1878
|
SSR
|
71.274 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TAG24_x_GPBD4_a - AhV
|
GM1878
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_Tifrunner_x_GT-C20_b - LG19
|
GM1878
|
SSR
|
19.5 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - A05
|
GM1878.A05
|
SSR
|
54.65 (cM)
|
Automated name-based
|
View Maps
|
|
seq8D9.A09
|
SSR
|
2.18
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - B10
|
seq8D9.B10
|
SSR
|
166.99 (cM)
|
Automated name-based
|
View Maps
|
|
PM346
|
SSR
|
2.28
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
PM346
|
SSR
|
29.864 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TAG24_x_ICGV86031_a - LG_AhXIII
|
PM346
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiPandey2012_a - LG_AhXIII
|
PM346
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
gi-1107.A09
|
SSR
|
3.49
|
peanut - AA_A.duranensis_x_A.stenosperma_d - A09
|
gi-1107
|
SSR
|
80.7 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.stenosperma_a - A09
|
gi-1107
|
SSR
|
9.8 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - B09
|
gi-1107.B09
|
SSR
|
50.64 (cM)
|
Automated name-based
|
View Maps
|
|
RN35H04
|
SSR
|
13.39
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TA09
|
RN35H04
|
SSR
|
60.497 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
RN35H04
|
SSR
|
51.414 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
PM170
|
SSR
|
13.89
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
PM170
|
SSR
|
49.361 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.stenosperma_a - A09
|
PM170
|
SSR
|
24.6 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
RN25B01
|
SSR
|
21.09
|
peanut - AA_A.duranensis_x_A.stenosperma_d - A05
|
RN25B01
|
SSR
|
75.6 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
RN25B01
|
SSR
|
59.114 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
ahFAD2A
|
CAPS
|
22.92
|
peanut - TT_Tifrunner_x_GT-C20_a - LGT9
|
ahFAD2A
|
CAPS
|
4.7 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AC3C07.A09
|
SSR
|
29.02
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - B08
|
AC3C07.B08
|
SSR
|
115.96 (cM)
|
Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - B08
|
AC3C07
|
SSR
|
41.413 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
GM2450
|
SSR
|
37.72
|
peanut - TT_Tifrunner_x_GT-C20_b - LG18
|
GM2450
|
SSR
|
32.3 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
GM2450
|
SSR
|
72.013 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
TC5D06
|
SSR
|
37.72
|
peanut - TT_AhComposite_SujayGowda2012_a - AhXIII
|
TC5D06
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiPandey2012_a - LG_AhXIII
|
TC5D06
|
SSR
|
69.56 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_Tifrunner_x_GT-C20_b - LG18
|
Ah1TC5D06
|
SSR
|
17.9 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TAG24_x_GPBD4_a - AhXIII
|
TC5D06
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TAG24_x_ICGV86031_a - LG_AhXIII
|
TC5D06
|
SSR
|
33 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_ICGS76_x_CSMG84-1_a - LG13
|
TC5D06
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
TC5D06
|
SSR
|
69.269 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.stenosperma_d - A05
|
TC5D06
|
SSR
|
75.6 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_ICGS44_x_ICGS76_a - LG12
|
TC5D06
|
SSR
|
50.6 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TA09
|
TC5D06
|
SSR
|
42.096 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_VG9514_x_TAG24_a - A09
|
TC5D06
|
SSR
|
40.7 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TG26_x_GPBD4_a - AhXIII
|
TC5D06
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
GM849
|
SSR
|
40.62
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
GM849
|
SSR
|
74.92 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
seq4G2.A09
|
SSR
|
40.66
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - B09
|
seq4G2.B09
|
SSR
|
78.82 (cM)
|
Automated name-based
|
View Maps
|
|
ARS768
|
SSR
|
46.68
|
No other positions |
|
seq9B4
|
SSR
|
48.56
|
peanut - AA_A.duranensis_x_A.stenosperma_d - A09
|
Seq9B4
|
SSR
|
24.5 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
seq16G8.A09
|
SSR
|
50.25
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - A01
|
seq16G8.A01
|
SSR
|
135.24 (cM)
|
Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.stenosperma_d - A01
|
Seq16G8
|
SSR
|
29.4 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
ARS786.A09
|
SSR
|
51.39
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - B08
|
ARS786.B08
|
SSR
|
115.36 (cM)
|
Automated name-based
|
View Maps
|
|
GM2359
|
SSR
|
54.49
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
GM2359
|
SSR
|
88.621 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_Tifrunner_x_GT-C20_b - LG18
|
GM2359
|
SSR
|
57.2 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.duranensis_a - A09
|
GM2359
|
SSR
|
52.557 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
Ah3TC25B04
|
SSR
|
54.58
|
No other positions |
|
ARS766
|
SSR
|
55.39
|
No other positions |
|
ARS752
|
SSR
|
56.59
|
No other positions |
|
GM1291
|
SSR
|
56.59
|
peanut - TT_ICGS44_x_ICGS76_a - LG12
|
GM1291
|
SSR
|
44.7 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
GM1291
|
SSR
|
88.798 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TG26_x_GPBD4_a - AhXX
|
GM1291
|
SSR
|
26.7 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiPandey2012_a - LG_AhXIII
|
GM1291
|
SSR
|
63.66 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.duranensis_a - A09
|
GM1291
|
SSR
|
57.258 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
ARS757
|
SSR
|
56.99
|
No other positions |
|
ARS759
|
SSR
|
56.99
|
No other positions |
|
GM2792
|
SSR
|
57.89
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A09
|
GM2792
|
SSR
|
84.144 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_Tifrunner_x_GT-C20_b - LG18
|
GM2792
|
SSR
|
49.8 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial