|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
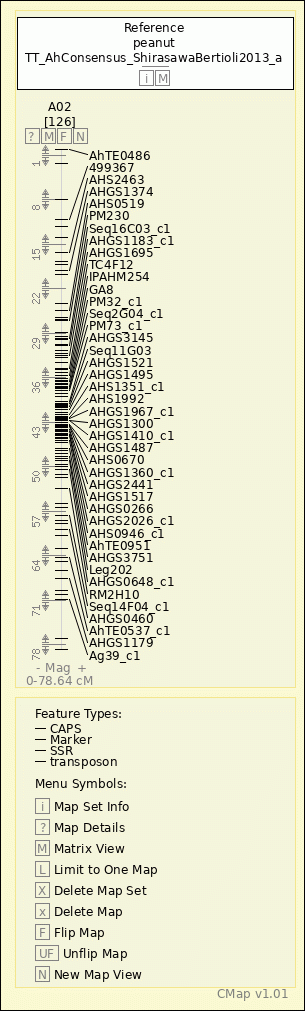
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - A02
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AhTE0486
|
transposon
|
0
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TA02
|
AhTE0486
|
transposon
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS2490_c1
|
SSR
|
2.216
|
No other positions |
|
AHGS1677_c1
|
SSR
|
7.966
|
No other positions |
|
499367
|
Marker
|
11.142
|
No other positions |
|
AHS0327
|
SSR
|
12.177
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TA02
|
AHS0327
|
SSR
|
23.964 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
498998
|
Marker
|
13.926
|
No other positions |
|
AHS2463
|
SSR
|
16.225
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TA02
|
AHS2463
|
SSR
|
15.918 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS1289_c1
|
SSR
|
17.772
|
No other positions |
|
TC42A05
|
SSR
|
18.145
|
peanut - AA_A.duranensis_x_A.stenosperma_b - AA02
|
TC42A05
|
SSR
|
0.409 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TA02
|
TC42A05
|
SSR
|
14.565 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.stenosperma_d - A02
|
TC42A05
|
SSR
|
10.7 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS1374
|
SSR
|
19.167
|
No other positions |
|
Seq11H01_c1
|
SSR
|
19.737
|
No other positions |
|
AhTE0465
|
transposon
|
24.351
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TA02
|
AhTE0465
|
transposon
|
33.907 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHS0519
|
SSR
|
25.416
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TA02
|
AHS0519
|
SSR
|
34.402 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
RGA18
|
Marker
|
26.496
|
No other positions |
|
AHS0314_c1
|
SSR
|
26.842
|
No other positions |
|
PM230
|
SSR
|
26.945
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - A02
|
PM230
|
SSR
|
47.54 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.stenosperma_a - A02
|
PM230
|
SSR
|
201.5 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS1940_c1
|
SSR
|
28.801
|
No other positions |
|
AHGS0655
|
SSR
|
29.549
|
No other positions |
|
Seq16C03_c1
|
SSR
|
29.808
|
No other positions |
|
RN20C10_c1
|
SSR
|
30.007
|
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - B02
|
RN20C10_c2
|
SSR
|
50.144 (cM)
|
Automated name-based
|
View Maps
|
|
AhTE0837
|
transposon
|
30.737
|
No other positions |
|
AHGS1183_c1
|
SSR
|
30.912
|
No other positions |
|
AHGS2345
|
SSR
|
31.672
|
peanut - AA_A.duranensis_x_A.stenosperma_b - AA02
|
AHGS2345
|
SSR
|
12.044 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
GM695
|
SSR
|
32.168
|
peanut - TT_AhConsensus_GautamiPandey2012_a - LG_AhII
|
GM695
|
SSR
|
48.6 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_ICGS76_x_CSMG84-1_a - LG14
|
GM695
|
SSR
|
0 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - A02
|
GM695
|
SSR
|
43.99 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_ICGS44_x_ICGS76_a - LG02
|
GM695
|
SSR
|
4.8 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_Tifrunner_x_GT-C20_b - LG15
|
GA166
|
SSR
|
27.7 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS1695
|
SSR
|
32.405
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial