|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
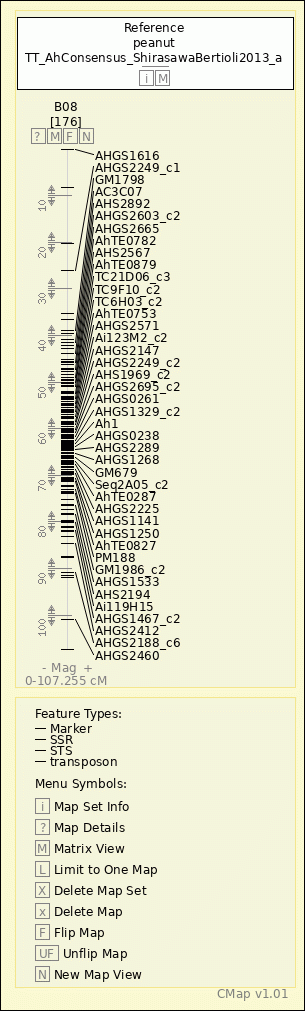
peanut - TT_AhConsensus_ShirasawaBertioli2013_a - B08
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
AHGS1616
|
SSR
|
0
|
No other positions |
|
AhTE0619
|
transposon
|
8.243
|
No other positions |
|
AhTE0006_c2
|
transposon
|
20.268
|
No other positions |
|
AHGS2249_c1
|
SSR
|
26.163
|
No other positions |
|
AhTE0846
|
transposon
|
35.317
|
No other positions |
|
AHGS1286_c2
|
SSR
|
36.48
|
No other positions |
|
GM1798
|
SSR
|
39.012
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - B08
|
GM1798
|
SSR
|
120.93 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.duranensis_a - A08
|
GM1798
|
SSR
|
59.613 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_Tifrunner_x_GT-C20_b - LG4
|
GM1798
|
SSR
|
57.8 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AhTE0724
|
transposon
|
39.592
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TB08
|
AhTE0724
|
transposon
|
9.968 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS1383_c2
|
SSR
|
40.85
|
No other positions |
|
AC3C07
|
SSR
|
41.413
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - A09
|
AC3C07.A09
|
SSR
|
29.02 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - B08
|
AC3C07.B08
|
SSR
|
115.96 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
Ai120I21
|
SSR
|
41.592
|
No other positions |
|
AHS0227_c2
|
SSR
|
42.06
|
No other positions |
|
AhTE0974
|
transposon
|
42.877
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TB08
|
AhTE0974
|
transposon
|
15.97 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHS2892
|
SSR
|
43.919
|
peanut - BB_A.ipaensis_x_A.magna_b - B08
|
AHS2892
|
SSR
|
77.7 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - BB_A.ipaensis_x_A.magna_a - BB08
|
AHS2892
|
SSR
|
10.621 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHS2968_c2
|
SSR
|
45.058
|
No other positions |
|
AHGS1470_c2
|
SSR
|
45.502
|
No other positions |
|
AHGS2603_c2
|
SSR
|
45.767
|
No other positions |
|
TC20B05_c2
|
SSR
|
46.198
|
No other positions |
|
AhTE0705
|
transposon
|
47.141
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TB08
|
AhTE0705
|
transposon
|
11.54 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS2665
|
SSR
|
47.264
|
peanut - BB_A.ipaensis_x_A.magna_a - BB08
|
AHGS2665
|
SSR
|
11.478 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
GM1760
|
SSR
|
47.775
|
peanut - TT_TG26_x_GPBD4_a - AhVIII
|
GM1760
|
SSR
|
77.9 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_Tifrunner_x_GT-C20_b - LG4
|
GM1760
|
SSR
|
41.2 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhComposite_SujayGowda2012_a - AhVIII
|
GM1760
|
SSR
|
38.669 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - A08
|
GM1760.A08
|
SSR
|
67.42 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - AA_A.duranensis_x_A.duranensis_a - A03
|
GM1760
|
SSR
|
13.955 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_TAG24_x_GPBD4_a - AhVIII
|
GM1760
|
SSR
|
50.5 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
peanut - TT_AhConsensus_GautamiFonceka2012_a - B08
|
GM1760.B08
|
SSR
|
23.1 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS1444
|
SSR
|
48.304
|
No other positions |
|
AhTE0782
|
transposon
|
48.96
|
No other positions |
|
AHGS1920
|
SSR
|
49.083
|
peanut - TT_RunnerIAC886_x_A.ipaensis-A.duranensis_a - TB08
|
AHGS1920
|
SSR
|
13.105 (cM)
|
Automated name-based, Automated name-based
|
View Maps
|
|
AHGS1186_c2
|
SSR
|
49.406
|
No other positions |
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial