|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
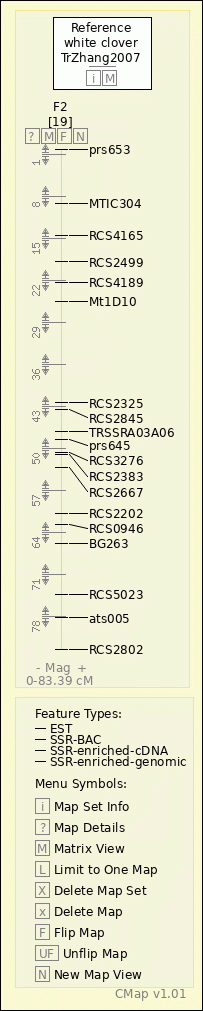
white clover - TrZhang2007 - F2
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
MTIC304
|
EST
|
9.07
|
No other positions |
|
RCS2499
|
SSR-enriched-cDNA
|
18.82
|
white clover - TrZhang2007 - E1
|
RCS2499
|
SSR-enriched-cDNA
|
56.72 (cM)
|
Automated name-based
|
View Maps
|
|
Mt1D10
|
SSR-BAC
|
25.36
|
No other positions |
|
RCS2845
|
SSR-enriched-cDNA
|
43.48
|
No other positions |
|
prs645
|
EST
|
48.39
|
white clover - TrZhang2007 - F1
|
prs645
|
EST
|
83.67 (cM)
|
Automated name-based
|
View Maps
|
|
RCS2383
|
SSR-enriched-genomic
|
50.87
|
white clover - TrZhang2007 - F1
|
RCS2383
|
SSR-enriched-genomic
|
10.13 (cM)
|
Automated name-based
|
View Maps
|
|
RCS2202
|
SSR-enriched-genomic
|
60.77
|
white clover - TrZhang2007 - C1
|
RCS2202
|
SSR-enriched-genomic
|
20.95 (cM)
|
Automated name-based
|
View Maps
|
|
BG263
|
EST
|
65.79
|
No other positions |
|
ats005
|
SSR-enriched-genomic
|
78.17
|
No other positions |
|
prs653
|
EST
|
0
|
No other positions |
|
RCS4165
|
EST
|
14.38
|
No other positions |
|
RCS4189
|
EST
|
22.25
|
white clover - TrZhang2007 - G1
|
RCS4189
|
EST
|
8.02 (cM)
|
Automated name-based
|
View Maps
|
|
RCS2325
|
SSR-enriched-genomic
|
42.33
|
No other positions |
|
TRSSRA03A06
|
SSR-enriched-genomic
|
47.16
|
No other positions |
|
RCS3276
|
SSR-enriched-genomic
|
50.57
|
No other positions |
|
RCS2667
|
SSR-enriched-genomic
|
53.12
|
No other positions |
|
RCS0946
|
SSR-enriched-genomic
|
62.56
|
No other positions |
|
RCS5023
|
EST
|
74.3
|
white clover - TrZhang2007 - F1
|
RCS5023
|
EST
|
115.71 (cM)
|
Automated name-based
|
View Maps
|
|
RCS2802
|
SSR-enriched-cDNA
|
83.39
|
white clover - TrZhang2007 - F1
|
RCS2802
|
SSR-enriched-cDNA
|
109.77 (cM)
|
Automated name-based
|
View Maps
|
 | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial  | CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial
| CMap Home | Maps | Map Search | Feature Search | Matrix | Map Sets | Feature Types | Map Types | Evidence Types | Species | Help | Tutorial